Fiber photometry enables researchers to visualize the activity of neuron populations in a freely-behaving animal using calcium imaging (Kim et al. 2016). By ‘reading out’ neural activity, fiber photometry data can be correlated with cognition and behaviour (e.g., the mouse pushes a lever and a neuron population fires). However, to determine a causal link between neural activity and behaviour, you must be able to manipulate (activate or inhibit) neural activity. The introduction of optogenetics has provided a technique to manipulate cell-type specific neural activity with high temporal precision using light (Yizhar et al. 2011) and, as a result, researchers can objectively determine if a neural population is linked with behaviour and cognition. Thus, the integration of optogenetics and fiber photometry can enable an all-optical approach to both ‘read’ and ‘write’ neural activity in a freely-behaving animal (Kim et al. 2017).
Can optogenetics be integrated with fiber photometry?
Biological Components
A wide-range of optogenetic probes have been developed, differing in function (excitation or inhibition), activation time, and expression properties.
Optogenetic probes are excited by wavelengths of light ranging from blue to red, depending on their properties. For example, Channelrhodopsin (ChR2) has been the optogenetic tool of choice for excitation of neural activity and halorhodopsin for inhibition due to their extensive development and use in the field of optogenetics.
Optogenetic probes have only an excitation spectrum to activate their excitation/inhibitory properties (Yizhar et al. 2011). In contrast, genetically encoded calcium indicators (GECI) behave in the same manner as a fluorophore (e.g. GFP), such that they exhibit excitation and emission spectra. However, the fluorescent signals of GECIs are dependent on intracellular calcium concentrations (i.e. increased fluorescence emission due to increased calcium influx), and these signals are capable of displaying dynamic behaviour, unlike a static signal emitted from a fluorophore (Grienberger & Konnerth 2012).
Researchers commonly propose the use of GCaMP for imaging and channelrhodopsin (ChR2) for optogenetics in all-optical experiments due to the optimization, efficiency, and frequent use of these biological probes.
However, a problem occurs with this combination: GCaMP and ChR2 have overlapping excitation spectrums (both with peak wavelength at ~470nm). When GCaMP and ChR2 are expressed in the same tissue, calcium imaging light excitation can potentially activate the optogenetic probe as well. Thus, GCaMP imaging and ChR2 optogenetic stimulation cannot be performed simultaneously due to potential optical crosstalk. Consequently, it is not possible to determine if the measured changes in the GCaMP signal are due to natural changes in neural activity or optogenetic-induced changes.
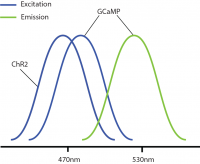
Optical crosstalk in all-optical experiments can be reduced by selecting imaging indicators and optogenetic probes with non-overlapping excitation spectrums (Emiliani et al. 2015).
Examples of combinations with reduced optical crosstalk are blue excitation/green emission imaging (e.g. GCaMP) and red-shifted optogenetics (e.g. Chrimson, Jaws) or green excitation/red emission imaging (e.g. RCaMP) and blue-shifted optogenetics (e.g. ChR2, GtAChR). Although there may be some overlap between the excitation spectrums of these probes, the likelihood of crosstalk will be reduced, preventing indirect activation of your optogenetic construct during imaging.
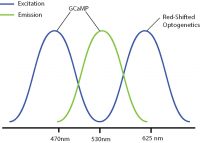
The further optimization of imaging and optogenetics probes (e.g. sensitivity, excitation spectra) will help prevent optical crosstalk in all-optical optogenetics and calcium imaging experiments.
Equipment Components
Optogenetics can be integrated into fiber photometry experiments, and the only additional components required are a filter set and the appropriate LED for optogenetic stimulation to combine with the calcium imaging LED. An example includes adding a red LED for red-shift optogenetics and GCaMP imaging (Kim et al. 2016).
Similar to calcium imaging with fiber photometry, optogenetic stimulation in such a system can only be performed widefield by stimulating all neurons within the field of view with no cellular resolution.
A key feature of fiber photometry is the possibility to perform multi-region optogenetics in combination with multi-region calcium imaging; however, in this setup, due to technical limitations, optogenetic stimulation cannot usually be conducted in one select region at a time and, instead, all regions will be stimulated simultaneously (Kim et al. 2016).
In conclusion, fiber photometry is a relatively easy-to-implement method to perform population all-optical calcium imaging and optogenetics experiments in one or multiple brain regions.