Published on 2025/03/17 Research powered by Mightex’s Polygon1000 

Yang, L., Zhu, A., Aman, J. M., Denberg, D., Kilwein, M. D., Marmion, R. A., … & Shvartsman, S. Y., ERK synchronizes embryonic cleavages in Drosophila. Developmental Cell, 59(23), 3061-3071 (2024).


The extracellular-signal-regulated kinase (ERK) pathway controls numerous developmental processes across species and has been linked to various human diseases, including neurocognitive disorders and cancers. In a recent study examining ERK signaling in Drosophila embryonic development, Liu Yang and colleagues employed optogenetic perturbations combined with phosphoproteomics to uncover a previously unknown function of ERK activation at embryonic poles. While ERK signaling has been extensively studied in developmental contexts, this study reveals its critical role in maintaining the speed and synchrony of embryonic cleavages – a finding that might have been missed using traditional genetic approaches alone.
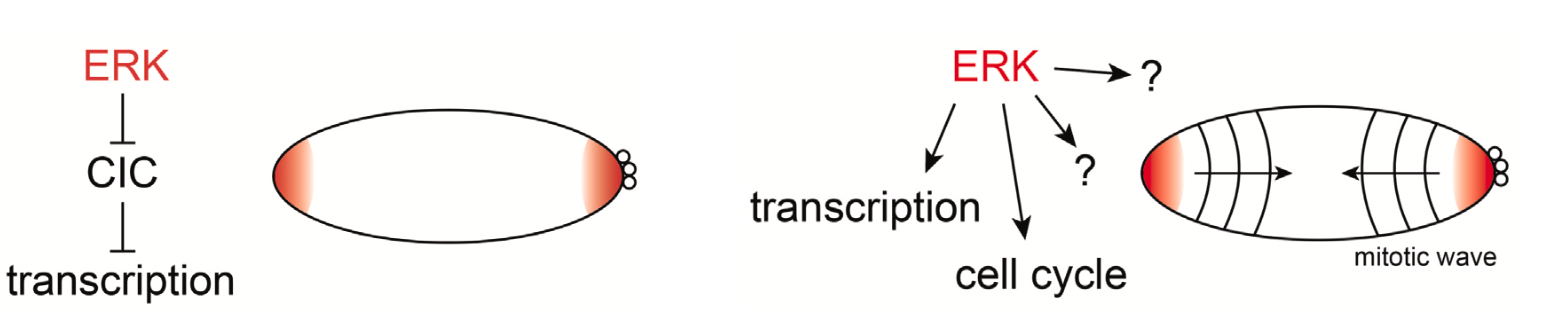
Figure 1: The previous (left) and revised (right) models for ERK activation in Drosophila
Drosophila embryos undergo rapid mitotic divisions during early development, with 13 successive division cycles completing within 2 hours of fertilization. While the first nine cleavages are fast and synchronous, the last four divisions are characterized by progressive slowing and loss of synchrony, with mitotic entry starting at the embryonic poles and spreading toward the middle [1]. This deceleration has been linked to activation of DNA damage checkpoint pathway [2], but the cause of advanced mitotic entry at the poles has remained elusive despite being noted in early studies of embryonic cleavages [3].
Methods and Results
To investigate ERK signalling in embryonic cleavages, the researchers used an optogenetic approach called optoSos to determine if ERK activation could directly trigger cell division [4], introducing this system into embryos that had fluorescently-labeled chromatin for visualization. The Mightex Polygon digital micromirror device (DMD) was used to achieve precise spatiotemporal control of ERK pathway activation. Imaging was performed on a Nikon A1 RS confocal microscope. For experiments, a 20 mm-wide stripe was illuminated using a 450-nm blue light for 0.1s every 6 seconds, synchronized with imaging using a 561-nm laser.
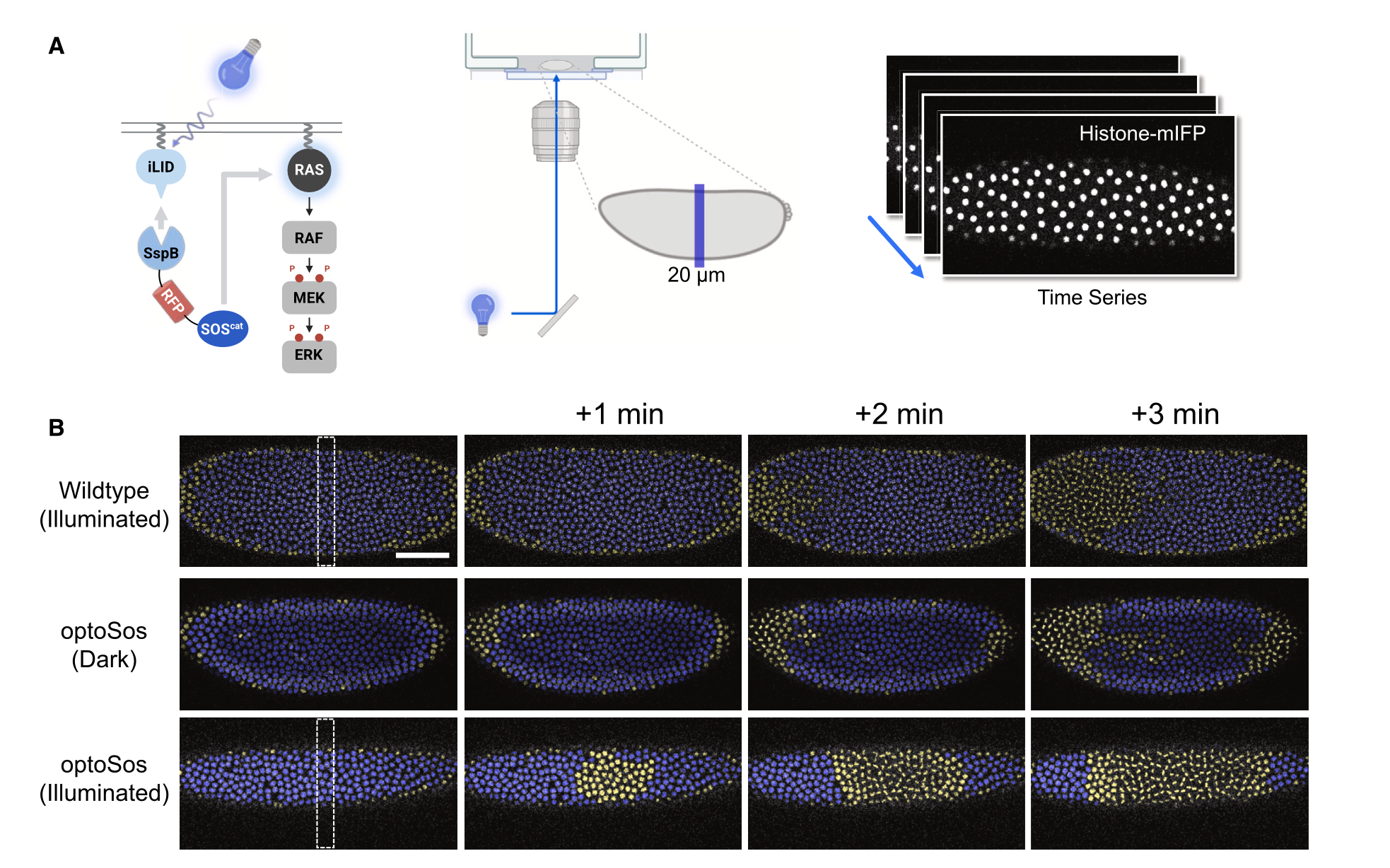
Figure 2: (A) Shows a schematic of the optogenetic system for localized ERK activation in a 20-mm wide stripe using the Mightex Polygon DMD. (B) Shows time-lapse images of wild-type and optoSos Drosophila embryos. Interphase and M-phase nuclei are represented in yellow and blue, respectively. The dotted box highlights the illuminated stripe of 20mm width
The results revealed that targeted optogenetic activation of ERK triggered mitotic entry, demonstrating that localized ERK signaling is sufficient to control cell cycle progression. This was particularly evident during the last two cleavage cycles, where the induced ERK activation generated a distinct pattern of mitotic entry, differing from normal embryonic development.
In summary, ERK signaling in embryos extends beyond controlling genes for larval terminal structures, affecting the entire embryo through direct substrates and phosphorylation cascades, with one confirmed function being the synchronization of embryonic cleavages. Understanding phosphorylation networks remains challenging due to their complexity, but the authors propose that analyzing responses to acute perturbations via optogenetics can help establish causal relationships and reveal how these networks respond to external signals and mutations. The Mightex Polygon DMD is instrumental for this approach, allowing targeted spatial and temporal control for optogenetic activation.
References
1. Di Talia, S., and Vergassola, M. (2022). Waves in Embryonic Development. Annu. Rev. Biophys. 51, 327–353. https://doi.org/10.1146/annurev-biophys-111521-102500.
2. Su, T.T., Campbell, S.D., and O’Farrell, P.H. (1999). Drosophila grapes/ CHK1 mutants are defective in cyclin proteolysis and coordination of mitotic events. Curr. Biol. 9, 919–922. https://doi.org/10.1016/s0960- 9822(99)80399-6.
3. Farrell, J.A., and O’Farrell, P.H. (2014). From egg to gastrula: how the cell cycle is remodeled during the Drosophila mid-blastula transition. Annu. Rev. Genet. 48, 269–294. https://www.annualreviews.org/content/journals/10.1146/annurev-genet-111212-133531
4. Toettcher, J.E., Weiner, O.D., and Lim, W.A. (2013). Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell 155, 1422–1434. https://doi.org/10.1016/j.cell.2013.11.004.
To read the full publication, please click here.



