Spatial control of microtubule dynamics at the subcellular scale by patterned illumination
Jeffrey van Haren & Torsten Wittmann
Department of Cell & Tissue Biology, University of California, San Francisco
Our lab focuses on elucidating the molecular mechanisms that control the organization of the mammalian microtubule (MT) cytoskeleton, a network of highly dynamic tube shaped polymers inside all of our cells that facilitates intracellular transport, controls chromosome segregation, serves to organize the cell interior, and allows cells to establish and maintain polarity. Proper organization of MTs inside cells requires precise spatial and temporal control of MT assembly/disassembly, but it has been difficult to study how MT regulatory factors accomplish such precise local control.
To gain a better understanding of these mechanisms, we have developed novel optogenetics tools that enable precise spatiotemporal control of the microtubule cytoskeleton with blue light. In collaboration with the Hahn lab at UNC, we devised a strategy to optically control proteins, by introducing a small light sensitive protein pair into a protein of interest. We used this strategy to develop a Photo-Inactivated variant of the key microtubule regulator EB1 (termed π-EB1), and generated cells in which π-EB1 replaced the wildtype protein (See figure 1A and B). Illuminating these π-EB1 cells with blue light, caused the nearly instant inactivation of π-EB1. Due to the rapid reversibility of this system, turning off the blue light resulted in rapid re-activation of π-EB1 and thereby allowed us to spatially control π-EB1 in small regions of the cell using the Polygon400 patterned illuminator (Mightex) (See figure 1C and D). This in turn allowed us to spatially control microtubule dynamics within cells and to study the immediate consequences of locally altering MT growth dynamics. Local inactivation of pi-EB1 in migrating cells caused cells to directly turn away from the area of π-EB1 inactivation. A manuscript describing our findings was recently published (van Haren et al. 2018, Nature Cell Biology).
This technique is likely applicable to many other proteins, and will be of great value to gain a better understanding of the mechanisms that control cell polarization. Current work is focused on elucidating local functions of EB1 during neuronal development.
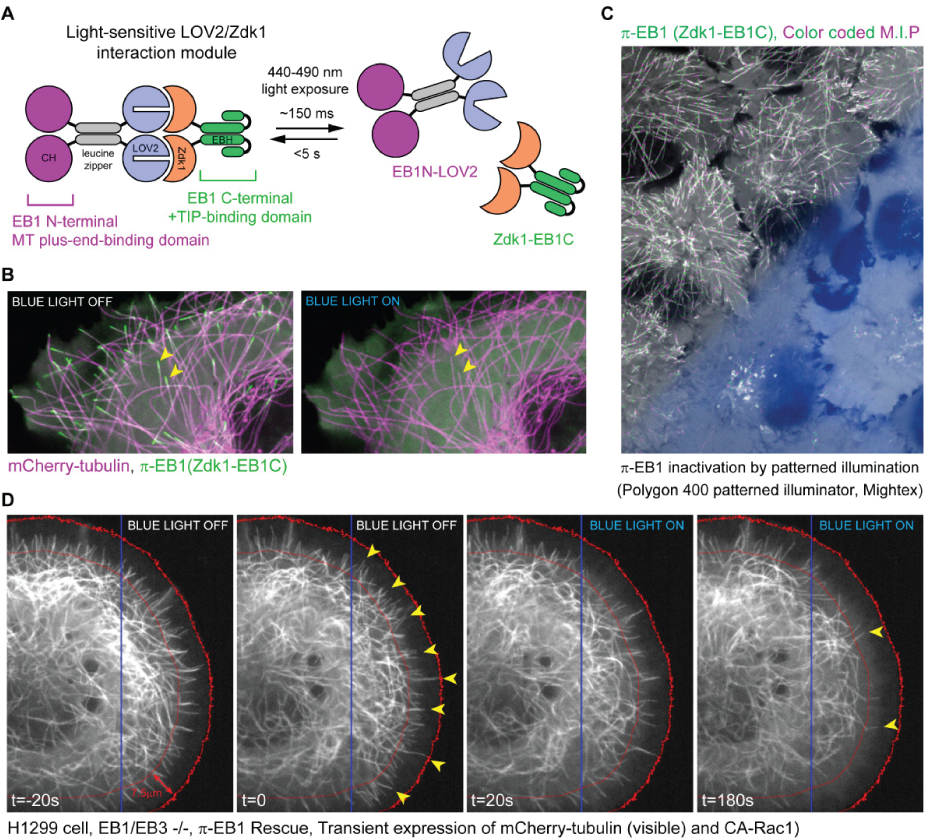
Figure 1 a) Schematic of the photo-inactivated π-EB1 design resulting in the reversible dissociation of the MT binding and +TIP adapter domains of EB1 upon blue light exposure. b) Cell expressing mCherry-tubulin (Microtubules), GFP-Zdk1-EB1C and unlabeled EB1N-LOV2 before and after blue light induced π-EB1 dissociation. Yellow arrowheads mark two microtubule ends before and after π-EB1 inactivation, and clearly show the loss of the GFP-Zdk1-EB1C from microtubule ends after blue light illumination c) Color-coded maximum intensity projection of a time lapse recording of cells expressing π-EB1. The lower half of the field of view was exposed to blue light using a digital micromirror device (Polygon 400, Mightex). Note the absence of EB1 comet tracks in the illuminated area, indicating that π-EB1 was locally inactivated d) π-EB1 cell expressing mCherry-tubulin and constitutively active Rac1, before and after π-EB1 inactivation in the right half of the cell (blue box marks area illuminated by the Polygon). Yellow arrowheads mark microtubules close to the cell edge (red lines mark a ~7.5 micron wide zone at the cell edge). Note the striking reduction of MTs at the cell edge after blue light inactivation (t=180s) of π-EB1(right panel).
Author: Jeffrey van Haren, Erasmus MC
Bio: Jeffrey van Haren received his PhD in cell biology from the Erasmus MC, where he studied the regulation of the mammalian microtubule cytoskeleton. He did his postdoctoral training in the Wittmann lab at UCSF, where he developed novel tools to control microtubule dynamics with light. In 2019 he started his own research group at the Erasmus MC.