
Spatiotemporal control of inflammatory lytic cell death through optogenetic induction of RIPK3 oligomerization
Published on 2024/07/08 Research powered by Mightex’s Polygon1000 
 Oh, T. J., Krishnamurthy, V., Han, J. W., Zhu, J., Beg, Z., Mehfooz, A., … & Zhang, K., Spatiotemporal control of inflammatory lytic cell death through optogenetic induction of RIPK3 oligomerization, Journal of Molecular Biology, 168628 (2024).
Oh, T. J., Krishnamurthy, V., Han, J. W., Zhu, J., Beg, Z., Mehfooz, A., … & Zhang, K., Spatiotemporal control of inflammatory lytic cell death through optogenetic induction of RIPK3 oligomerization, Journal of Molecular Biology, 168628 (2024).

Introduction
The ability of our bodies to maintain cellular stability requires continual and programmed cell death. Various mechanisms control how cells die and serve different functions within their tissue environments. One such process is necroptosis, which differs from more commonly known apoptotic or necrotic cell deaths as marked by cytokine production and the cell’s plasma membrane rupture. While evidence suggests necroptosis is beneficial in viral infections, it is also a pathogenic marker for many diseases. Studying the signaling pathways for necroptosis may elucidate our understanding of the development and treatment of such diseases.
Necroptosis is usually initialized by the activation of known signaling pathways, which necessitates the binding of a ligand to a receptor to catalyze the signaling cascade. Tumor necrosis factor alpha (TNFa) is a well-established ligand that initiates necroptosis by binding its receptor, TNF receptor 1 (TNFR1) which eventually through multiple subsequent events leads to the activation of Receptor Interacting Protein Kinase 3 (RIPK3), an essential enzyme in both canonical and non-canonical necroptosis. Here, the authors developed a light-activatable RIPK3 protein (La-RIPK3) that can be optogenetically activated (Figure 1). This allows precise spatiotemporal control of necroptotic pathways bypassing ligand activation and enabling the further dissection of this pathway.
Methods and Results
La-RIPK3 was designed by incorporating a Cryptochrome2 (CRY2) oligomerization domain with mCherry-tagged RIPK3. Cells transduced with La-RIPK3 that were exposed to blue light had significantly increased cell death as detected by the SytoxGreen marker. Blue-light-exposed La-RIPK3 expressing cells also showed increased expression of cytokines and rupture of the cell membrane, supporting the activation of necroptotic pathways.
One of the benefits of having a light-activatable system is the ability to spatially control the activation of the signaling pathway. In this publication, this was achieved by using the Mightex Polygon1000 DMD illuminator to project blue light only in targeted ROIs within the field of view (FOV) (Figure 2). Specifically, the authors divided the FOV into 4 quadrants using Mightex PolyScan software and only illuminated 2 diagonally opposed quadrants. Only cells within the 2 illuminated quadrants were detected with the SytoxGreen marker, indicating that necroptosis pathways were only activated there. Cell morphology assays also supported this finding. Spatially defined activation of necroptosis may prove beneficial for future studies in larger cellular contexts, including tissue morphology and model organism development.
The development of optogenetic tools such as La-RIPK3 is especially helpful in the dissecting of signaling pathway spatiotemporal dynamics. Not only do they advance our knowledge of molecular mechanisms, but when applied to different contexts, they can be great assets to dissect further cellular, developmental, and pathogenic processes.
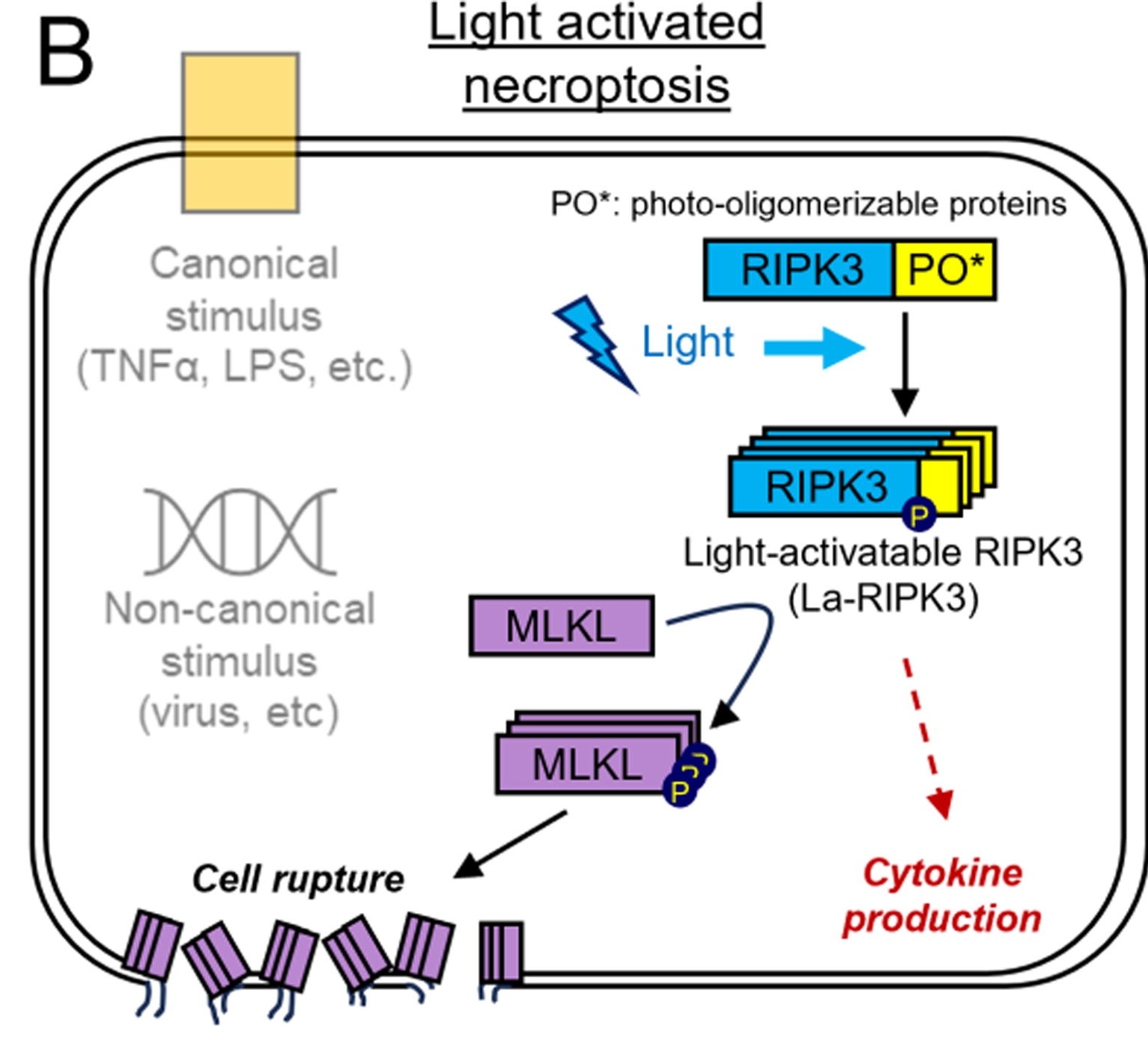
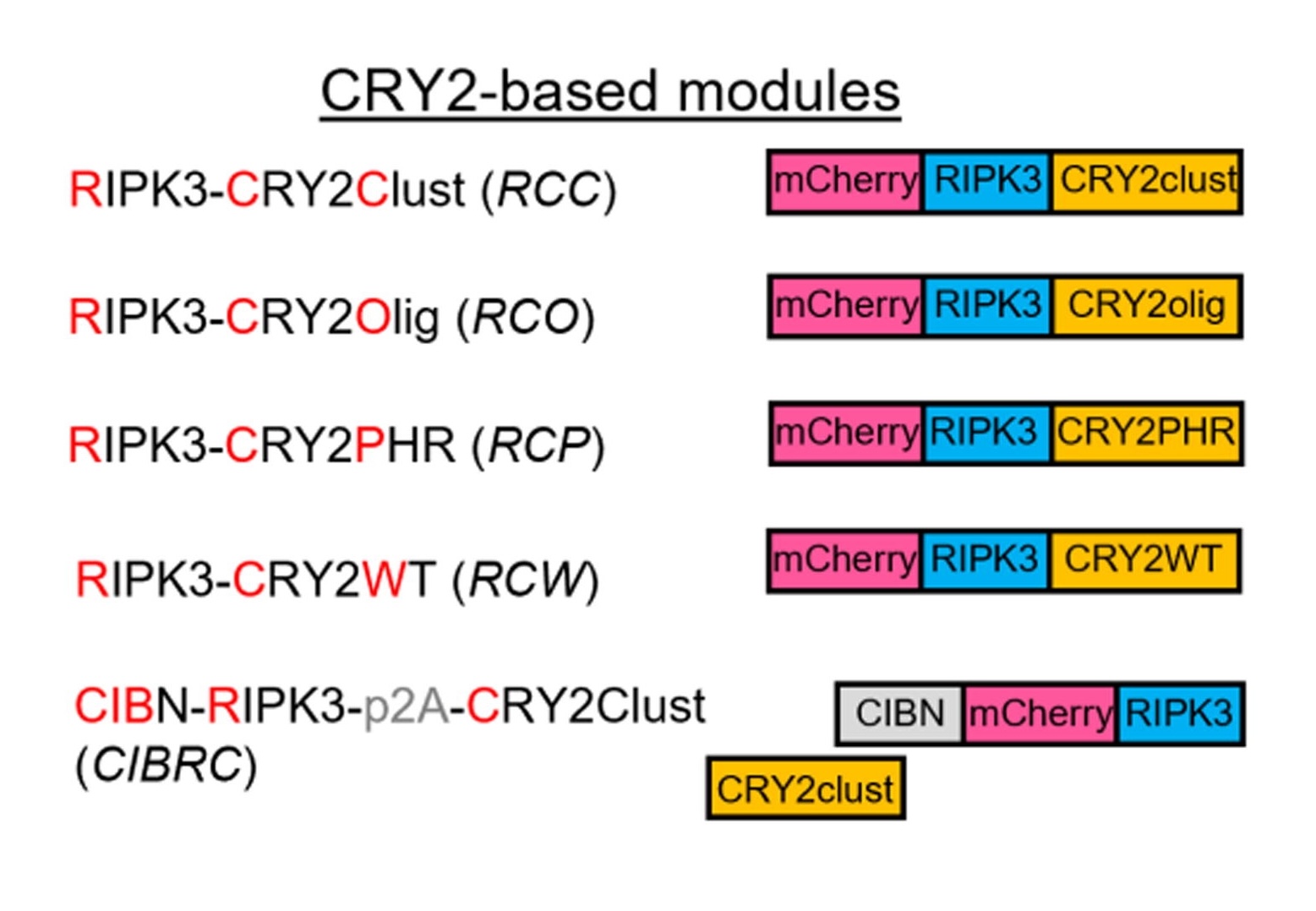
Figure 1: La-RIPK3 design and necroptosis pathway.
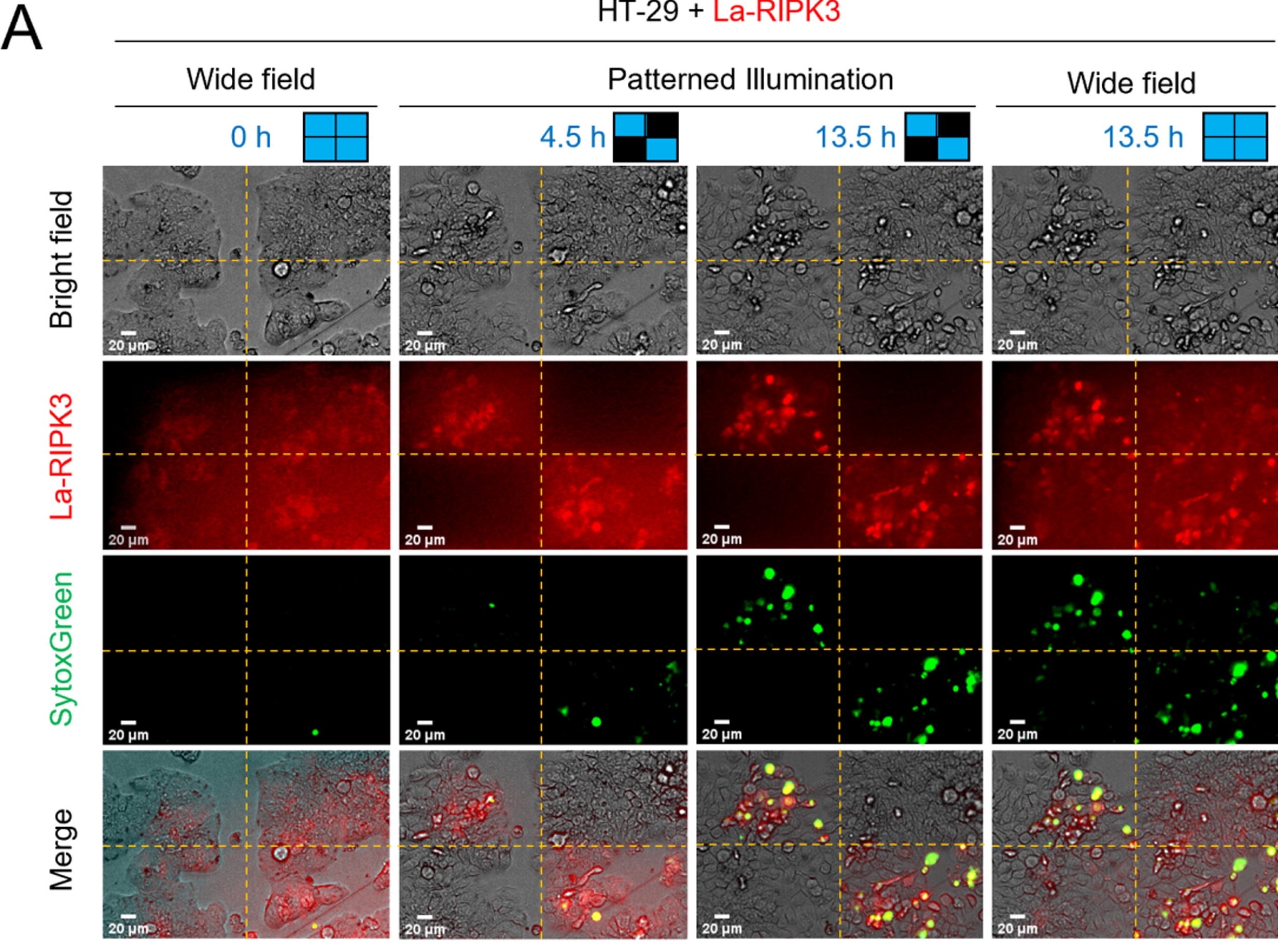
Figure 2: Spatially defined activation of necroptosis using Mightex Polygon1000 DMD illuminator. Only La-RIPK3 expressing cells in illuminated quadrants are induced into necroptotic pathways as indicated by the SytoxGreen labeling.
Francisco Rodrigues, PhD Manager, Customer Solutions at Mightex
To read the full publication, please click here.